
Mammal, Bacteria, and Plant Cell Membrane Biophysics
Introduction
The cell membrane is the cornerstone of the work we do. As scientists, we are curious about how it works, its function, and the role it plays in life; but as engineers, we are interested in leveraging what we learn to develop new biotechnologies.
To begin, a cell membrane is a thin film, only several nanometers thick, that is made up of lipids and proteins. Lipids are molecules that are amphiphilic, meaning they have one end that loves water and one end that hates water. Because of this characteristic, these molecules self assemble into a “bilayer” structure, or essentially two rows of lipids that align with the water-loving ends outside and the water-hating ends inside. It looks something like this:

Proteins are biomolecules that are often embedded in the lipid bilayer and are designed by nature to carry out specific functions. You might think of these molecules and the sensory organs of the cell that help the cell assess and understand its environment and communicate with other cells. Proteins can span across the lipid bilayer, making them “transmembrane proteins” or be associated with the membrane in a variety of other ways, and so then referred to as “peripheral proteins.” Some proteins bind to specific species on the membrane. When you add the proteins to the membrane, it looks something like to this:
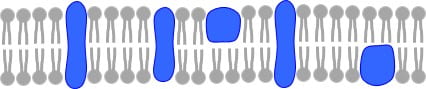
Another important class of molecules are glycans, which are sugar groups that decorate lipids and proteins to give them distinct characteristics and binding abilities. Sugar groups help proteins fold properly and govern its interactions with other proteins and cell surfaces. These sugar groups help to distinguish plant cells from bacteria or mammal cells. Cholesterol is also an important component of many cell membranes. Cholesterol is a kind of “membrane buffering agent” that keeps the fluidity of the membrane in a range necessary so biochemical processes occur normally. Cholesterol also fosters the formation of “lipid domains” that are thought to play special roles sequestering apart, or coalescing together, specific biomolecules to orchestrate when they react. Putting this complexity in, the cell membrane may look something like this:
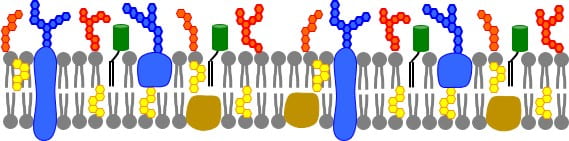
In our research, we are interested in the functions of the cell membrane as a unit and of the species within it, often referred to as the “biophysics” of the cell membrane. We are fascinated by the membrane’s ability to orchestrate the many reactions necessary for life and its complexity. Thus, we examine the mechanical properties of membranes, the diffusion behavior of the lipids and proteins comprising it, and phenomena that occur when the cell surface interacts with external species.
Background: Methodologies
In our work, we use advanced microscopy techniques to track single protein and lipids in membranes as they move about. These motions give us information about the local environment of the protein or lipid and an overall sense of the heterogeneity of the membrane.
We used quartz crystal microbalance with dissipation to study the mechanical properties of membranes. This technique relies on an oscillating crystal that changes resonance when mass is added. With the technique we can determine when material binds to a membrane and if that material disrupts it. This powerful technique gives us real-time information about changes to the membrane properties and integrity.
We make extensive use of microfluidic devices in our work, using the Cornell Nanofabrication Facility to make our prototype devices. By combining microfluidic devices with supported biomembranes, we create renditions of the cell surface that we use to carry out our studies and develop new technologies.
The Daniel Lab Innovations
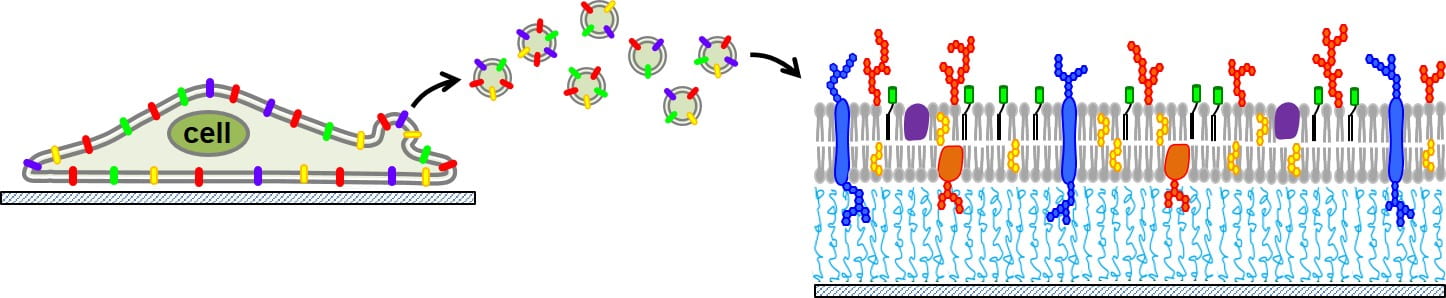
We often study cell membranes in the form of a supported lipid bilayer (SLB). This is a convenient format because SLBs are planar, self-assemble in microfluidic channels, and are compatible with various kinds of microscopy and surface techniques. However, they are typically made by lipid reconstitution. The Daniel lab made a huge leap forward by developing a process to extract vesicles from cell membranes and assemble them onto planar supports without having to disrupt the compositions and complexity of the native cell. We use a process called “blebbing” to initiate the cell to release vesicles from its plasma membrane surface, collect them, and rupture these vesicles into planar sheets on a supporting surface. These molecularity-complex SLBs contain the lipids, proteins, glycans, and cholesterol found in the native cell and remain functional and active. Our team has succeeding in making these bilayers from plant, bacteria, and mammal cells. We have been able to put them on a variety of surfaces as well, including conducting polymers, which forms the basis of the bioelectronics work we do. Here is an idea of how this process works:
What the Daniel Lab is Working On Now
- Host membrane mimics for pathogen interaction studies
- Diffusion and confinement of transmembrane proteins in membranes using single molecule tracking
- Biomembrane chips for sequential bioenzymatic reactions
- New dual-mode biomembrane biosensing devices
- Leaf on a chip
- Discovery of the Rules of Life