
Host-Pathogen Interactions
Background: Virus Infection

A virus can be pictured as a capsule filled with genetic material. This capsule is decorated with outwardly protruding spikes. These spikes pierce the host cell membrane and pass genetic material inside the cell. This genetic material infects the cell, disrupts its function, and initiates disease.
The ability of a virus to invade a cell is governed by two primary interactions, both of which are guided by the viral spike proteins. First, the virus must bind to the cell surface. Second, the virus membrane must merge with the host cell membrane, a process called membrane fusion. The success of these steps depends on the chemical groups on the virus spikes, the chemical groups on the host cell surface, and local chemical environment in which the virus and host cell meet.
Background: Methodologies
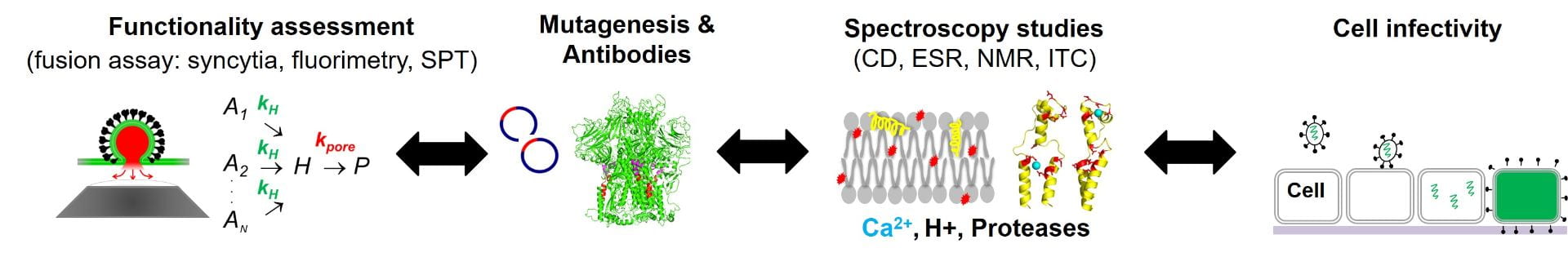
We bring a distinctly “engineering ” approach to studies of virus entry and infection. The studies we conduct make use of biophysical, structural, and biochemical tools as well as techniques from molecular biology. This work is highly interdisciplinary and we work closely with researchers from Microbiology & Immunology, Chemistry, and Bioechemistry on these projects. The figure below illustrates our approach to this research and how it integrates together.
The Daniel Lab Innovations
With current techniques, it is simple to study a virus binding to the cell surface. However, studying fusion is more complicated. A virus and cell can be proximate, yet not fused. Visual observation of the virus shell merging with the cell surface to form a fusion pore is obscured by size and proximity of both components. Thus, one must look for a secondary signal to report the occurrence of fusion. Additionally, activation of fusion occurs when cues from the cell or surrounding tissue are received by the virus. These cues vary with cell type and environment.
Certain cues make a healthy cell susceptible to viral infection. However, the extent to which these cues influence the ability of a virus shell to fuse with a specific host cell type is unknown. The Daniel Lab developed a technique which allows the study of viral fusion with host cells while controlling the cues to which the viruses are exposed. This approach makes it possible to test conditions, such as the cellular difference between species, in order to identify which conditions impact virus fusion.
These are time-lapse, single particle tracking images made using the Daniel Lab technique. The images show virus fusion triggered by proton uncaging to rapidly drop the local pH. Each fluorescence burst marks a fusing virus.

What We’re Working On Now
The Daniel Lab determines which environmental cues enhance virus fusion to various cell types, and identifies mutations in the virus spike which increase fusion using single particle tracking. We carry out these studies with models for the following viruses:
- Influenza, which has annual outbreaks in both hemispheres.
- Coronavirus, which gave rise to SARS in 2003 and MERS in 2012.
- Ebola, which has had outbreaks as recently as 2019.
Matching mutations to fusion highlights chemical groups which lead to increased rates of infection. We examine different chemical groups on the host cell which act as receptors for viruses. From this, we determine which interactions between the virus and cell favor fusion.
Why This Research Matters
Identifying factors which control fusion, and understanding this step in the infection cycle contributes to development of both anti-viral medication and syn-viruses, or virus-like nanomachines. Syn-viruses can be used for therapeutic drug delivery or local cell environmental control and signaling.